Our analytical methods are modular and can be profitably combined.
Select one of our services to learn more about the method.
Our mission is to answer your questions.
The polymerase chain reaction (PCR) is the most fundamental and important working method of IDENTXX.
Four components are needed to perform a PCR successfully
PCR procedure
For qualitative detection, also known as end-point PCR, the enzyme reaction is first carried out completely. After the end of the reaction, the resulting products are separated according to size in an agarose gel and made visible with the help of fluorescent dyes.
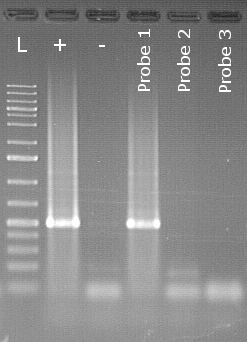
Specific endpoint PCR detection of Peronospora belbahrii
The example shows an agarose gel of a detection of Peronospora belbahrii downy mildew of basil from seeds. Next to the size marker (L) is the positive control (+) followed by the negative control (-). In the lane of the positive control you can see a clearly white shining area, which is also called the "band". This is the stained specific PCR product in the lane of sample 1 there is also a band at the same height. The samples are therefore assessed as positive. Samples 2 and 3 do not show any bands and are therefore evaluated as negative.
Our qualitative PCR detections are safe and robust, and are used in the detection of a wide variety of pathogens.
Another important PCR method is quantitative PCR (qPCR), which is also called real-time PCR. It follows the same reaction principles as qualitative PCR. However, the reaction solution is supplemented by another component, the so-called probe. This is located between the primers and is also specific for the DNA section being searched for.
If a PCR reaction now takes place, the polymerase enzyme degrades the probe. A fluorescent dye (fluorophore) released in the process can be detected in real time during this degradation process. The more PCR products are produced, the more fluorophore is released. The thermocycler displays this measured value. The earlier there is an exponential increase in the curve, the more DNA was present in the initial solution.
The use of probes has another advantage. By using different fluorophores, we can perform up to 5 PCR reactions in one reaction tube (multiplex analyses). This makes it possible to set the content of specific pathogen DNA (shown in blue in the upper curve) in relation to the DNA of the host plant (shown in green).
This approach has several advantages:
We also used endpoint PCR as a basis for DNA sequencing, distinguishing between Sanger sequencing and pyrosequencing. These are two important analytical tools that we used for different analytical purposes.
This method was developed by Frederick Sanger and Alan Coulson in the 1970s and, along with the PCR reaction, forms the foundation of modern molecular biology. The method is as simple as it is ingenious. Special oligonucleotides, which are labelled with fluorescent dyes, are added to the finished endpoint PCR reaction solution. In a subsequent PCR reaction, DNA fragments of different lengths are obtained, each of which carries a fluorescence-labelled oligonucleotide at the end, which can be analysed with the help of appropriate detectors. The result is a sequence of coloured peaks with the corresponding assignment to the four bases of the DNA.
An endpoint PCR serves as the basis for pyrosequencing. By using a biotinylated primer, one strand of each of the resulting PCR products carries biotin at the end (labelling). Through covalent binding to streptavidin, this labelled DNA strand can be separated and purified. During the sequencing reaction, the four bases are added separately to the reaction solution. If an insertion takes place, an enzyme reaction-based light flash is released, which is detected in real time as in qPCR.
The crucial point is the quantification of the nucleotide incorporation. This allows the degree of zygosity of a TSR position to be determined exactly.
An endpoint PCR serves as the basis for pyrosequencing. By using a biotinylated primer, each strand of the resulting PCR product carries biotin at the end (labeling). Through the covalent binding to streptavidin, the labeled DNA strand can be separated and cleaned. During the sequencing reaction, the four bases are added separately to the reaction solution. If incorporation takes place, an enzyme reaction-based flash of light is released, which is detected in real time, as in qPCR.
The crucial point is the quantification of the nucleotide incorporation. This allows the degree of zygosity of a TSR position to be determined exactly.
Endpoint PCR serves as the basis for fragment length analysis. By using a fluorophore-labelled primer, each strand of the resulting PCR products carries a fluorescent dye (labelling) at the end. With the help of high-resolution capillary electrophoresis, which is equipped with a detector unit for fluorescence signals, the lengths of the PCR fragments can be determined to the nucleotide.
A fragment length analysis is used in the following analyses:
To identify methylated cytosines, the DNA must be subjected to a bisulphite reaction before the endpoint PCR. In this reaction, an uracil is inserted for all cytosines that are not methylated and are directly in front of a guanine base (CpG dinucleotide).
In the subsequent endpoint PCR, the uracil is replaced by thymine, resulting in a base sequence that differs from the original.
The sequence can be determined after the end of the reaction either by the Sanger method or by the Pyro method.
A methylation analysis is used in the following applications:
In cooperation with two leading institutions, IDENTXX offers greenhouse biotesting to determine metabolic resistance, also known as nTSR.
The partners have both a modern spray stand and sufficient greenhouse capacity and specialist staff.
The following points are particularly worth mentoning:
Contact
Address
Information